The limitation of phenotypic taxonomy, which is based on the ability of morphological characteristics to distinguish species, often leads to incorrect identification, especially for cryptic species, which require high levels of expertise (Vega-Sánchez et al., 2020 ![]() ). In recent decades, molecular taxonomy has helped to resolve the "classification crisis", during which many scientists have suggested the use of phylogenetic taxonomy instead of morphological identification (Kozlov et al., 2016
). In recent decades, molecular taxonomy has helped to resolve the "classification crisis", during which many scientists have suggested the use of phylogenetic taxonomy instead of morphological identification (Kozlov et al., 2016 ![]() ).
).
Odonata are a highly diverse group of aquatic insects. This order is primitively divided into two suborders, Anisoptera and Zygoptera. They are predatory insects that help control harmful aquatic organisms. During their larval stage (called naiads), they spend most of their time in various freshwater environments, such as shallow ponds, swamps, and rivers. When they reach adulthood, they transform into flying terrestrial insects with colors that vary from bright to pale. They are often found near wet areas, and some species are migratory (Attaullaha et al., 2021 ![]() ).
).
DNA barcoding is one of the most common taxonomic systems that has been particularly successful in diagnosing and identifying new species from different groups (Koroiva et al., 2022 ![]() ). Identification via this method, which relies on sequence information from short fragments of primary DNA sequences, can be performed simply and affordably, without the need for a taxonomist in the group.
). Identification via this method, which relies on sequence information from short fragments of primary DNA sequences, can be performed simply and affordably, without the need for a taxonomist in the group.
In addition, the success of DNA-based species identification strategies can provide comprehensive information about the geographic and genetic diversity of the population of interest (Gaytán-Pinzón et al., 2022 ![]() ).
).
The DNA sequences of each species are unique; they can be viewed as "barcodes" and are used to resolve misidentifications, which may cause taxonomic inconsistencies inherent in the type of taxonomy practiced to date in reference databases (Salvi et al., 2020 ![]() ). Several genes, including those encoding the mitochondrial protein COXI and 16S and 28S rRNA, have been selected for use in animal barcoding studies because they possess a wide range of phylogenetic signals and exhibit rapid nucleotide substitution rates, which help distinguish between cryptic species and reveal phylogenetic structures within a species (Jackson et al., 2014
). Several genes, including those encoding the mitochondrial protein COXI and 16S and 28S rRNA, have been selected for use in animal barcoding studies because they possess a wide range of phylogenetic signals and exhibit rapid nucleotide substitution rates, which help distinguish between cryptic species and reveal phylogenetic structures within a species (Jackson et al., 2014 ![]() ; Luo et al., 2022
; Luo et al., 2022 ![]() ).
).
The COI universal primers are very powerful, allowing the construction of demonstrative sequences for most animal groups (Kim et al., 2014 ![]() ), whereas Abdullah et al. (2024
), whereas Abdullah et al. (2024 ![]() ) reported that the use of specific primers provides more precise and reliable quantification results where the design of appropriate target-specific primers avoids primer matches with nontarget species and allows undesired amplification.
) reported that the use of specific primers provides more precise and reliable quantification results where the design of appropriate target-specific primers avoids primer matches with nontarget species and allows undesired amplification.
Previous diagnostic studies did not survey entire genera of Odonata, were based on a small number of characters or did not use established phylogenetic methods (Bybee et al., 2021 ![]() ). Recently, the evolutionary relationships of the Odonata group have been studied on a molecular basis, with increasing emphasis on molecular genetics and even the lowest ordinal level (Dijkstra et al., 2014
). Recently, the evolutionary relationships of the Odonata group have been studied on a molecular basis, with increasing emphasis on molecular genetics and even the lowest ordinal level (Dijkstra et al., 2014 ![]() ). However, the lack of large-scale studies based on high-throughput data that can be linked to an evolutionary perspective has prevented comparative questions about the evolutionary levels of these insects (Letsch and Gottsberger, 2016
). However, the lack of large-scale studies based on high-throughput data that can be linked to an evolutionary perspective has prevented comparative questions about the evolutionary levels of these insects (Letsch and Gottsberger, 2016 ![]() ).
).
Libellulidae is the most species-rich family within Anisoptera, with more than 900 described species distributed worldwide and found in all aquatic habitats and biota. The incidence of sexual dimorphism is high in this family because it adapts physiologically and morphologically during its life cycle to facing unsuitable conditions in the environment (Casas et al., 2018 ![]() ).
).
Some local studies have addressed the taxonomy and ecology of Odonata adults of several Anisoptera and Zygoptera species, such as Ali and Khidhir (2015 ![]() ), who studied Odonata adults in northern Iraq, and Abd and Al-Asady (2014
), who studied Odonata adults in northern Iraq, and Abd and Al-Asady (2014 ![]() ). Al-Hashmi (2017
). Al-Hashmi (2017 ![]() ) studied central and southwestern Iraq; however, for naiads of some Odonata species, there are morphological identification studies by Darweesh (2018
) studied central and southwestern Iraq; however, for naiads of some Odonata species, there are morphological identification studies by Darweesh (2018 ![]() ) and Ahmed and Kareem (2019
) and Ahmed and Kareem (2019 ![]() ; 2020
; 2020 ![]() ; 2024
; 2024 ![]() ), who studied the morphological identification of Odonata adults and nymphs in Basrah Province, southern Iraq. However, no molecular studies on this insect group of Iraq have been recorded in public databases such as BOLD, with the exception of one study by Geraci et al. (2011
), who studied the morphological identification of Odonata adults and nymphs in Basrah Province, southern Iraq. However, no molecular studies on this insect group of Iraq have been recorded in public databases such as BOLD, with the exception of one study by Geraci et al. (2011 ![]() ), who documented information about the genetic sequences of some Odonata species in the marshes of Iraq at the National Center for Biotechnology Information (NCBI). Our study aimed to obtain accurate identification and more detailed confirmation of the molecular genetics of some endemic species of Anisoptera from Basrah Province as a base step for documenting the DNA sequence information of the Odonata mtCOXI gene in the national GenBank via the BOLD system and DNA barcodes.
), who documented information about the genetic sequences of some Odonata species in the marshes of Iraq at the National Center for Biotechnology Information (NCBI). Our study aimed to obtain accurate identification and more detailed confirmation of the molecular genetics of some endemic species of Anisoptera from Basrah Province as a base step for documenting the DNA sequence information of the Odonata mtCOXI gene in the national GenBank via the BOLD system and DNA barcodes.
2. Materials and Methods
2.1 Ethical approval
No ethical approval is required for this study.
2.2 Sampling
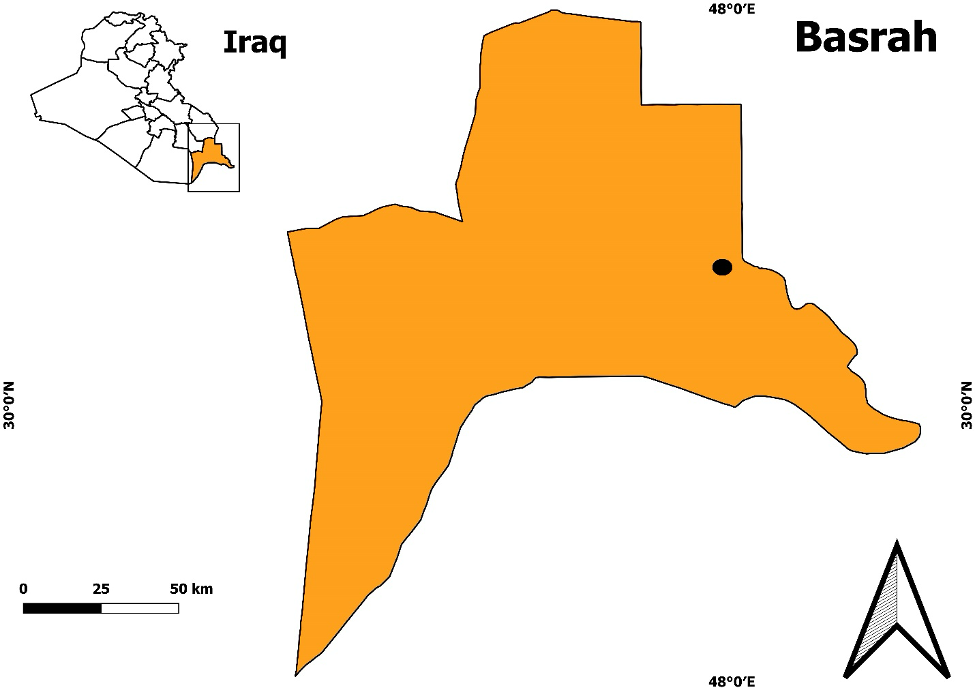
Samples of 50 individuals of dragonflies (order Odonata) belonging to 5 species were collected from wetland regions with geographical extents of 30°25 45.1'N–47°55 52.1'E in Basrah Province, southern Iraq, between May and December 2018 (Figure 1). Dragonfly adult samples were collected randomly from the field by hand or with aerial nets, whereas the nymphs were collected via an aquatic insect net or sieve (30 cm diameter and 1 mm mesh size) and preserved by freezing at -15°C after each species was placed in a separate collection bottle with the assigned date of collection and location (Ahmed and Kareem, 2019). Morphological identification was performed according to Degabriele (2013 ![]() ).
).
2.3 Primer design
Species-specific primers were designed by downloading sequences from selected species of Odonata on the National Center for Biotechnology Information (NCBI) website to choose nucleotide-based arrangements of mitochondrial COI gene markers (Lim et al., 2011 ![]() ). The downloaded sequences are then imported into the BioEdit application; to perform multiple alignments on the acquired target and nontarget sequences, the ClustalW option in the BioEdit application was chosen, and pieces of 144--327 bp nucleotide bases are then entered into the Primer3Plus website to obtain some recommendations for forward and reverse primer pairs to be used.
). The downloaded sequences are then imported into the BioEdit application; to perform multiple alignments on the acquired target and nontarget sequences, the ClustalW option in the BioEdit application was chosen, and pieces of 144--327 bp nucleotide bases are then entered into the Primer3Plus website to obtain some recommendations for forward and reverse primer pairs to be used.
We found that the optimum primer pair can be determined through criteria based on the %GC and Tm (melting temperature) values, where the web tool of NCBI's Basic Local Alignment Search Tool (BLAST) is the best option for performing primer specification validation, as summarized in Table 1.
Table 1. Details of selected species of Dragonflies, target genes and nucleotide sequence primers used in this study.
2.4 DNA extraction and amplification
Mitochondrial DNA was dissected following the manufacturer's instructions for a DNA extraction kit (gSYNC™ DNA extraction kit; Geneaid, Taiwan). From frozen samples of selected dragonflies, the thoracic and legs of adults and some nymphs were dried with liquid nitrogen and then crushed or ground as much as possible via a ceramic mortar; the powder was lifted to remove the remaining coarse parts. Each sample weighed 0.20 grams and was placed into a 1.5 ml microcentrifuge tube. The sample genetic material was then isolated or stored at -20°C if not used immediately. The extraction results revealed that the concentrations of the genetic material ranged from 6.2--58 ng/µl and that the purity ranged from 1.73-1.98 (Table 2).
Table 2. Concentration and purity of the genetic material (DNA) extracted via NanoDrop.
2.5 Molecular analysis, PCR and sequencing
In accordance with the methods of Lee et al. (2012 ![]() ), the DNA samples were analyzed via agarose gel (1%) electrophoresis and stained with ethidium bromide. Specialized primers were synthesized for each selected Anisoptera species from Basrah Province for the first time. The primers were prepared by the Bioneer Company as a dried product at different concentrations. The primers were dissolved in free water (DNase/RNase) to reach a final concentration equivalent to 100 pmol/microliter, and the stock solution was prepared at a concentration equivalent to 10 pmol/microliter for analysis. In addition, several experiments were conducted to obtain the Tm and Ta temperatures of the primers and to determine the optimal temperature that gave the best results via polymerase chain reaction (PCR).
), the DNA samples were analyzed via agarose gel (1%) electrophoresis and stained with ethidium bromide. Specialized primers were synthesized for each selected Anisoptera species from Basrah Province for the first time. The primers were prepared by the Bioneer Company as a dried product at different concentrations. The primers were dissolved in free water (DNase/RNase) to reach a final concentration equivalent to 100 pmol/microliter, and the stock solution was prepared at a concentration equivalent to 10 pmol/microliter for analysis. In addition, several experiments were conducted to obtain the Tm and Ta temperatures of the primers and to determine the optimal temperature that gave the best results via polymerase chain reaction (PCR).
The optimal concentration of the template DNA was determined by comparing the brightness of the amplified fragment with that of a band from a size marker (5000 bp) in agarose gel electrophoresis.
The amplified PCR products (20 μl for each species) were sequenced in both directions, forward and reverse, by using an automated sequencer Genetic Analyzer by Yang Ling Biotechnology Co. (China).
2.6 Phylogenetic identification
The PCR products were analyzed via Bioinformation BioEdit (software V.7.2.6, Network V.5, Mega 7.0), aligned and filtered, and then a Basic Local Alignment Search Tool (BLAST) search was performed on the online portal of the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov), the sequence matrices of the species were constructed for identity and homology of the analyzed sequences.
2.7 Molecular sequence products in GenBank
To obtain accession numbers from NCBI, representative sequences of the partial COI gene for each species of Anisoptera (nymph or adult) from Basrah Province were deposited for the first time in the DNA Data Bank of Japan (DJB) database (which is a biological database that collects DNA sequences located at the National Institute Genetics Institute in Shizuoka, Japan). It is a member of the International Nucleotide Sequences Collaborative Database (INSD) and exchanges its data with the European Biology Laboratory (EMBL) and the National Center for Biotechnology Information (NBCI) daily; thus, researchers can register and upload sequence data of their sample to GenBank at any of these centers, which will be saved in the global database.
Each DNA sequence of the diagnostic species from Basrah was matched with the recorded species in NCBI from different geographic regions of the world, the percentage of identity was determined via BLAST, and a phylogenetic tree was drawn. The evolutionary development of the species was compared with that of identical species in other geographical regions, and the genetic distance between them was calculated.
2.8 Barcodes of molecular sequence products
The DNA barcode images of the sequences submitted were developed via web-based https://ngdc.cncb.ac.cn/databasecommons/ database/id/4609 barcode at Insect Barcode Informatica (IBIn), NBAII database- India, by uploaded to the Barcode of Life Data (http://www.boldsystems.org). The required information for submission of the barcode of the target gene includes the order, family, and species of the insect for which the barcoding has been performed; the longitude and latitude of the location of the collected samples; the country; the barcode marker (name of the gene); the source (reference); the author name and institute address; the nucleotide sequence; and the image of the insect. Once the submission of the barcode record was performed, a unique barcode image was automatically created. Immediately after the submission of the barcode record, e-mail alerts are sent automatically to the experts through database administrative members.
3. Results
The five species of Anisoptera (Figure 2) that were morphologically identified as the species Hemianax ephippiger (Family: Aeshnidae), Diplacods trivialis, Crocothemis erythraea (naiad), Orthetrum sabina naiad, O. sabina adult and Trithemis annulata (Family: Libellulidae), and their scientific taxonomy are as follows,
Order: Odonata
Suborder: Anisoptera
Family: Aeshnidae
Genus: Hemianax Selys, 1883
- 1. Hemianax ephippiger Burmeister, 1883
Family: Libellulidae
Genus: Crocothemis Brauer 1868
- 2. Crocothemis erythraea (Brulle, 1832)
Genus: Diplacodes Kirby, 1889
- 3.D. trivialis (Rambur, 1842)
Genus: Orthetrum Newman, 1933
- 4.Orthetrum sabina (Drury, 1770)
Genus: Trithemis Brauer 1868
- 5. Trithemis annulata (Palisot de Beauvois, 1807)

The results of electrophoresis on agarose gels confirmed the success of amplifying the cytochrome oxidase mtCOXI gene via primers specific for each species of selected dragonflies, as the results revealed DNA bands with molecular weights ranging from 144--327 base pairs (Figure 3).

3.1 Genetic analysis
Details of the alignment and filtering of the genetic sequence via Maga7.0 are shown in Figure 4, which illustrates the results of the DNA sequence of the COXI gene and the number of nucleotide bases for each selected species of Anisoptera from Basrah Province, Iraq.
The molecular identities of five species, Crocothemis erythraea, Diplacodes trivialis, Orthetrum sabina, Trithemis annulata, and Hemianax ephippiger,were established, and the sequences were deposited in the NCBI database for the first time (Table 3).
Table 3. Similarity percentage of mtCOX1 for the selected species of Anisoptera from Basrah Province, with the accession numbers of similar species in the GenBank database.
The nucleotide sequences of the mtCOXI gene were subjected to BLAST, and DNA barcodes were successfully developed for selected Anisoptera species from Basrah. The results revealed 91.8% to 100% similarity to the NCBI database of Odonata species, and the molecular identity of our species was confirmed.
The total length of the product varied from species to species and ranged from 295-111 bp. The final analyzed sequences were submitted to GenBank with an accession number (Table 4).
3.2 Phylogenetic tree
The neighbor-joining (NJ) tree was inferred via MEGA 0.7 software to determine the evolutionary history, with evolutionary distances calculated via the composite maximum likelihood method (Ibragimov and Has' Minskii, 2013 ![]() ). Units of the number of base substitutions per site were measured per site.
). Units of the number of base substitutions per site were measured per site.
Table 4. The sequences of the Anisoptera species from Basrah Province were submitted to GenBank and IBIN.
The analysis involved five nucleotide sequences, and any ambiguous positions for each sequence pair were excluded. Figure 5 shows the evolutionary relationships between the species of selected odonates in the present study and their corresponding species in countries around the world. Two main branches were divided into five secondary branches. The first branch was divided into two branches. The first branch included the species Diplacodes trivialis from Iraq and the species D. nebulosa from Thailand. The second branch included two strains of the species Trithemis annulata from Iraq and Africa. The third branch isolated two strains of Crocothemis erythraea from Iraq and Namibia. The fourth branch included two strains of Orthetrum sabina from Iraq and Singapore. The fifth branch included Anax ephippiger from Namibia and its synonymous Hemianax ephippiger from Iraq. The genetic ratio between species in the branches of the evolutionary tree ranged from 0.000--0.98, with a scale bar of 0.05. Evolutionary analyses were conducted in MEGA 7 as previously described (Kumar et al., 2016 ![]() ).
).

3.3 DNA Barcodes
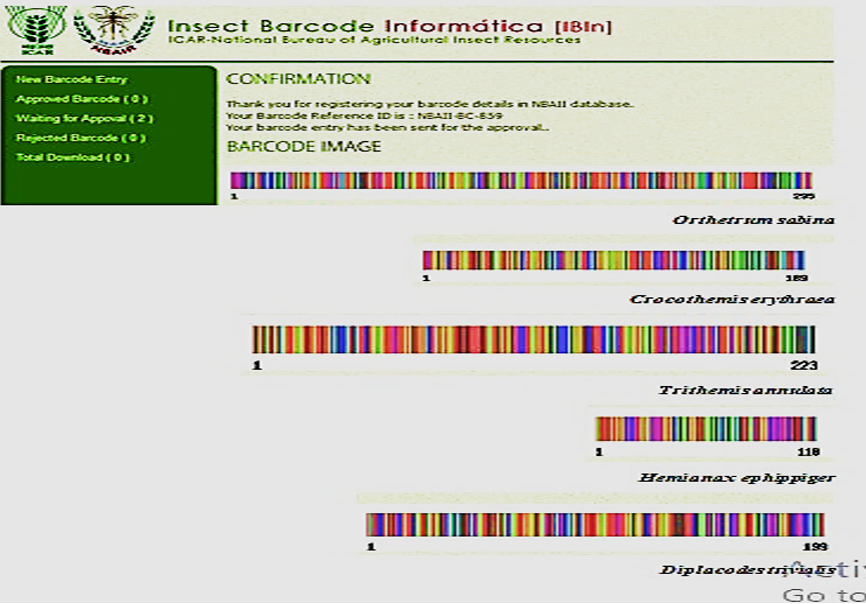
A barcode for the genetic sequence and a special reference number for each species were recorded in the NBAII database, as shown in Figure 6, on the basis of the documented information in GenBank.
4. Discussion
We designed a specific primer for each selected Anisoptera species from Basrah Province to provide more accurate and specific matches with target species. In agreement with recent molecular studies, this study revealed that the COX gene could help identify Odonata when it was used as the basis of the universal bioidentification system for animals at the regional scale (Jin et al., 2022 ![]() ; Koroiva et al., 2022
; Koroiva et al., 2022 ![]() ).
).
The sequences of the products in our study were of good quality when DNA from the thorax and legs of Anisoptera species was isolated to match the identities of the selected species via both morphological and molecular identification. Although most taxonomic studies depend on phenotypic or morphological characteristics to diagnose the species of the Odonata order, which includes approximately 6,000 described species, we have confidence that the most accurate and important probability justifiable identification lies in the use of DNA sequences as taxonomic barcodes.
DNA analysis methods provide important insights into the distribution of genetic diversity around the world (Zheng et al., 2023 ![]() ). Larvae of Libellulidae unknown by phylogenetic relationships can be identified with certainty and in a reliable manner (Huang et al., 2020
). Larvae of Libellulidae unknown by phylogenetic relationships can be identified with certainty and in a reliable manner (Huang et al., 2020 ![]() ); all the sequences and species were identified via the BLAST tool on NCBI, which is available on the BOLD website (www.barcodinglife.com), for morphological identification with DNA barcoding identification.
); all the sequences and species were identified via the BLAST tool on NCBI, which is available on the BOLD website (www.barcodinglife.com), for morphological identification with DNA barcoding identification.
In our study, we performed a BLAST search at NCBI to identify the specific species. For C. erythraea, the similarity was 100% with Namibia species, where this species is common worldwide, and this fact could be seen for O. sabina, which is 100% identical with the same species in Singapore. T. annulatawas present in 99.10% of the African population, whereas H. ephippiger, which belongs to the family Aeshnidae, was 100% identical to the same species from Japan and 99.06% identical to the species from Namibia.
However, we found a close match for Diplacodes trivialis, but the maximum genetic similarity was 91.8% with D. nebulosa from Thailand, and the e value was 0. These species are phenotypically very similar, but D. nebulosa has not been recorded in Iraq until now, and the ratio of genetic similarity may be due to the presence of some strains of this species in the Arabian region. Furthermore, our findings support that the NJ, BLAST, and BOLD methods are dependable for the analysis of DNA barcodes.
Our results of sequence analysis revealed that a high frequency of identical amino acids remained in the ClustalW results because the selected species belong to Odonat and Anisoptera, where the two families Libellulidae and Aeshnidae are closely related. To resolve the relationships among Odonata species morphologically, many researchers have attempted to identify different characteristics to identify odonates on the basis of wing venation and copulatory organs (Sacchi and Hardersen, 2013 ![]() ), but no one has reached considerable conclusions. Bybee et al. (2021
), but no one has reached considerable conclusions. Bybee et al. (2021 ![]() ) reported that the genomic data they obtained were able to resolve parts of the topology of the studied families of Odonata and shed light on possible evolutionary scenarios that may have shaped the evolutionary lineage of this taxon.
) reported that the genomic data they obtained were able to resolve parts of the topology of the studied families of Odonata and shed light on possible evolutionary scenarios that may have shaped the evolutionary lineage of this taxon.
The structure of the phylogenetic tree for the selected dragonfly species from the Basrah region revealed that the DNA sequences of Diplacodes trivialis and Trithemis annulata were highly similar and close, whereas those of Crocothemis servilia, Orthetrum sabina species and Hemianax ephippiger were more similar, although those of H. ephippiger from different families were similar. Therefore, all the selected species belong to the same suborder, which is related to the DND contents.
Close species with high levels of similarity sequences are almost close taxonomically, whereas divergence is shown in the distant taxa groups, which means that the genetic information and signals are different in diverse regions, as shown by (Kumar et al., 2016 ![]() ).
).
Our results are consistent with previous findings of Casas et al. (2018 ![]() ), who used the mitochondrial COXI gene to estimate genetic relationships between different members of the Libellulidae family. Additionally, many genetic studies have addressed the role of odonates in genetic diversity from an evolutionary point of view and morphological variation and confirmed the species diagnosis (Jisha Krishnan and Sebastian, 2015 a
), who used the mitochondrial COXI gene to estimate genetic relationships between different members of the Libellulidae family. Additionally, many genetic studies have addressed the role of odonates in genetic diversity from an evolutionary point of view and morphological variation and confirmed the species diagnosis (Jisha Krishnan and Sebastian, 2015 a ![]() ; b
; b ![]() ).
).
Finally, the identification via molecular data of these species was fully identical to that of the four species of Crocothemis erythraea, Diplacodes trivialis,Hemianax ephippiger, and Trithemis annulata, which commonly have a high level of dimorphism in body coloration between males and females, especially in the family Libellulidae, as mentioned by Corbet (1999 ![]() ). Therefore, the problem of misdiagnosis can be solved by molecular analysis, and this is what we observed through our study.
). Therefore, the problem of misdiagnosis can be solved by molecular analysis, and this is what we observed through our study.
5. Conclusions
The use of DNA barcoding has proven to be a valuable tool for identifying dragonfly species, especially in Libellidus. Our work represents an initiative for the molecular identification of dragonflies in Iraq, helping to accurately document the local fauna of Odonata in the national genus GenBanck.
Acknowledgements
The authors are grateful to Dr. Ali J. Al-hassona for laboratory assistance and the members of the Department of Biology, College of Pure Science, and the Marine Science Center, University of Basrah, for providing the laboratory facilities during the study.
Data availability statement
Not applicable.
Informed consent statement
Not applicable.
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
Sample collection, species identification, DNA extraction and write the manuscript: HKA and DKK. Supervision: DKK. All the authors critically reviewed the manuscript and agreed to submit a final version of the article.



