Life depends on water. Water supply has been essential to society for many uses, including drinking, agriculture, industry, domestic use, and more, from the beginning of human civilization. The lack of access to clean drinking water is a major cause of health issues in poor nations (Hasan et al., 2019 ![]() ; Bylund et al., 2017
; Bylund et al., 2017 ![]() ; Thayer et al., 2012
; Thayer et al., 2012 ![]() ). Health and disease are significantly impacted by water, sanitation, and hygiene. Without a question, access to clean water is essential for survival and cannot be achieved without it (Hutton and Chase, 2016
). Health and disease are significantly impacted by water, sanitation, and hygiene. Without a question, access to clean water is essential for survival and cannot be achieved without it (Hutton and Chase, 2016 ![]() ).
).
Several water sources in developing nations are dangerous due to the presence of dangerous chemical, biological, and physical contaminants (Cheesbrough, 1985 ![]() ). Infectious disorders that are spread by water are known as waterborne diseases because the pathogen or causal organism is present in the water and is consumed (Hurst, 2018
). Infectious disorders that are spread by water are known as waterborne diseases because the pathogen or causal organism is present in the water and is consumed (Hurst, 2018 ![]() ).
).
Water-borne infections, including diarrhea and gastrointestinal disorders, have been the source of many outbreaks. These illnesses are caused by various bacteria, viruses, and protozoa (Kristanti et al., 2022 ![]() ).
).
The traditional waterborne enteric pathogens are Shigella spp. (four species causing dysentery), Salmonella enterica (subsp. enterica ser. Typhi, causing typhoid), and Vibrio cholerae (sero-groups O1 and O139, causing cholera). These pathogens have been largely controlled by water treatment/disinfection, and as a result, they are rarely a problem via drinking water in developed regions. However, Shigella sonnei, along with closely related shiga toxin and verotoxin-producing E. coli persist in the sewage of developed nations due to person-to-person and foodborne transmission (Ashbolt et al., 2015 ![]() ).
).
A genus of rod-shaped, gram-negative, non-spore-forming enterobacteria, mostly motile, is called salmonella. They are facultative anaerobes and chemo organotrophs, deriving their energy from organic substances through oxidation and reduction reactions. Growing most species on media containing ferrous sulfate allows one to easily detect hydrogen sulfide that they create. There are two phases that most isolates go through: a motile phase (I) and a non-motile phase (II). It is possible to transition non-motile cultures into the motile phase after primary culture. Closely linked to the Escherichia genus, salmonella can be found in both warm- and cold-blooded animals, including humans, as well as in food-borne illnesses including typhoid fever and paratyphoid fever (Stella et al., 2018 ![]() ; Ryan and Ray, 2004
; Ryan and Ray, 2004 ![]() ).
).
Shigella are non-motile, facultative anaerobic, gram-negative rods that do not generate spores. Shigella's pathogenicity, physiology (inability to digest lactose or decarboxylate lysine), and serology set it apart from the closely related Escherichia coli bacterium. The most frequent cause of bacillary dysentery is Shigella. This illness is not the same as enterotoxigenic diarrhea caused by Escherichia coli, which results in excessive amounts of watery diarrhea. The genus contains four sero-groups: A with twelve serotypes of S. dysenteriae, B with six serotypes of S. flexneri, C with eighteen serotypes of S. boydii, and D with one serotype of S. sonnei (Strockbine et al., 2015 ![]() ).
).
Although E. coli is considered to be a more accurate indication of faecal pollution, it still has a number of drawbacks that should be taken into account before relying solely on the findings of E. coli testing for faecal contamination (Rana et al., 2024 ![]() ; Hossain et al., 2024
; Hossain et al., 2024 ![]() ; Li et al., 2021
; Li et al., 2021 ![]() ). It has even been demonstrated that E. coli can flourish in certain natural aquatic settings. The emergence of dangerous strains of Escherichia coli and their pervasiveness in sources of drinking water are concerning (Saima et al., 2021
). It has even been demonstrated that E. coli can flourish in certain natural aquatic settings. The emergence of dangerous strains of Escherichia coli and their pervasiveness in sources of drinking water are concerning (Saima et al., 2021 ![]() ). There is little evidence to correlate serotype to pathogenicity, and there is no biochemical marker to distinguish pathogenic from non-pathogenic strains of E. coli (Rivas et al., 2015
). There is little evidence to correlate serotype to pathogenicity, and there is no biochemical marker to distinguish pathogenic from non-pathogenic strains of E. coli (Rivas et al., 2015 ![]() ). E. coli strains that cause disease have been isolated from mountain streams, drinking water sources, and tap water. According to reports, one of the ways that pathogenic E. coli strains spread is by water contamination; this is the case for many outbreaks of the newly developing O157:H7 strain of enterohemorrhagic E. coli (Gebregziabher et al., 2024
). E. coli strains that cause disease have been isolated from mountain streams, drinking water sources, and tap water. According to reports, one of the ways that pathogenic E. coli strains spread is by water contamination; this is the case for many outbreaks of the newly developing O157:H7 strain of enterohemorrhagic E. coli (Gebregziabher et al., 2024 ![]() ; Ashbolt, 2015
; Ashbolt, 2015 ![]() ; Hruday et al., 2003
; Hruday et al., 2003 ![]() ).
).
The water-borne pathogens EPEC, ETEC, EIEC, and EHEC are well-known. It has a relatively limited lifespan outside of the environment, and it might not be able to thrive if there are other faecal bacteria present. In water, E. Coli O157:H7 can enter a viable but non-culturable (VBNC) condition but cannot live for extended periods of time. (Gambushe et al ., 2022 ![]() )
)
Vibrio are tiny, curved, Gram-negative rods that have a single polar flagellum. Facultative anaerobes with both respiratory and fermentative metabolisms are called vibrios (Rana et al., 2024 ![]() ). Sodium is an absolute need for most species and encourages growth in all. The structures known as fimbriae, or pili, found on Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus cells are composed of the protein TcpA. One of the main factors influencing in vivo colonization is TcpA production, which is co-regulated with the expression of the cholera toxin (Cabral, 2010
). Sodium is an absolute need for most species and encourages growth in all. The structures known as fimbriae, or pili, found on Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus cells are composed of the protein TcpA. One of the main factors influencing in vivo colonization is TcpA production, which is co-regulated with the expression of the cholera toxin (Cabral, 2010 ![]() ).
).
The World Health Organization has identified antibiotic resistance in bacteria as one of the biggest risks to human health (WHO, 2014 ![]() ). Antibiotic-resistant bacteria are able to multiply due to the selection pressure that is created when antibiotics are overused or misused (Islam et al ., 2024
). Antibiotic-resistant bacteria are able to multiply due to the selection pressure that is created when antibiotics are overused or misused (Islam et al ., 2024 ![]() ; Woappi et al., 2016
; Woappi et al., 2016 ![]() ; Levy et al., 2005
; Levy et al., 2005 ![]() ). The environmental contamination is thought to be the most effective method for both the selection of resistant populations and the transfer of resistance genes via mobile genetic elements (Larsson and Flach, 2022
). The environmental contamination is thought to be the most effective method for both the selection of resistant populations and the transfer of resistance genes via mobile genetic elements (Larsson and Flach, 2022 ![]() ; Ahmad et al., 2021
; Ahmad et al., 2021 ![]() ). This species may serve as an indicator of bacteria that spread antibiotic resistance in aquatic environments because of their great prevalence there and a variety of antibiotic-resistant mechanisms (Siri et al., 2023
). This species may serve as an indicator of bacteria that spread antibiotic resistance in aquatic environments because of their great prevalence there and a variety of antibiotic-resistant mechanisms (Siri et al., 2023 ![]() ). It may serve as a storehouse for ARGs that can spread horizontally through genes to other dangerous bacteria (Tao et al., 2022
). It may serve as a storehouse for ARGs that can spread horizontally through genes to other dangerous bacteria (Tao et al., 2022 ![]() ).
).
The current study was conducted to look into the occurrence of enteric pathogens (such as E. coli, Salmonella spp., Shigella spp., and Vibrio spp.) in various sources of water. Molecular characterization of isolated enteric pathogens, including the discovery of drug-resistant and virulence factor genes. Finally, an antibiogram research was conducted to choose appropriate antibiotics for therapeutic purposes, hence reducing economic loss.
2. Materials and Methods
2.1 Ethical approval
No ethical approval is required for this study.
2.2 Study areas
The entire study was carried out in the Department of Microbiology and Hygiene, Faculty of Veterinary Science, Bangladesh Agricultural University, Mymensingh-2202, Bangladesh, from March 2021 to October 2022. The places pond water samples were collected from the different ponds inside of Mymensingh city where as river water samples were collected from the Brahmaputra which flow beside Mymensingh city (Figure 1).

2.3 Sample collection and processing
A total of 40 water samples (pond water, tap water, tube-well water, and river water) were collected, among which were 10, 10, 10, and 10 samples from rivers, ponds, water taps, and tube-wells, respectively. Each type of sample was collected from five different places of Mymensingh city where 2 samples were collected from each of the five different locations (Table 1). All water samples of this study were carefully handled and kept in transport box and maintained the temperature at 40C and immediately after collection all samples transported to the department of microbiology and hygiene for bacterial analysis. Using sterile falcon tubes, the samples (100 ml each) were collected, and then they were carefully transferred to the proper sterile containers and brought to the laboratory for bacteriological investigation. The water that had been collected was centrifuged for 3 minutes at 10,000 rpm. Sediments were re-suspended in sterile PBS for culture, while supernatants were disposed of.
Table 1. Sample types, number of samples, and sampling location.
2.4 Isolation and identification of bacteria
After being re-suspended, the primary culture of the samples was inoculated with NB and incubated for an entire night at 37ºC. The enriched culture from nutrient broth was streaked on to selective agar media and incubated at 37ºC for 24 hours. To obtain pure cultures, a single colony that had surfaced on the selective media was streaked onto selective media. The standard procedure was followed when examining various samples culturally for bacteriological analysis (ICMSF, 1988 ![]() ). Based on colony morphology, Gram's staining reaction, and biochemical test, bacteria were identified. The pure culture was stained by Gram using the technique outlined by Cheesbrough (2006
). Based on colony morphology, Gram's staining reaction, and biochemical test, bacteria were identified. The pure culture was stained by Gram using the technique outlined by Cheesbrough (2006 ![]() ).
).
2.5 Molecular identification of Salmonella spp., Shigella spp., E. coli, and Vibrio spp. by PCR
Each isolate's pure bacterial colony was inoculated into NB and kept at 37°C for the overnight. Next, 1ml of cultured broth was centrifuged for 3 min at 12,000 rpm. The supernatant was disposed of and replaced with 200 µl of purified water. After that, the tube was placed in boiling water and left there for 20 min. It was then immediately placed on ice for a brief period of time roughly for 10 min and centrifuged for 10 min at 12,000 rpm. During PCR, the supernatant was collected and used as a DNA template. Master Mix for PCR 10 µl, 1 µl each of the forward and reverse primers, 4 µl of the DNA template, and 4 µl of nuclease-free water. The final final volume of PCR product was 20 µl. The primers were performed to amplify the DNA of bcfC, invC, 16S rRNA, and groEL genes of Salmonella spp., Shigella spp., E. coli, and Vibrio spp., respectively
Table 2. The sequence of primers for bacteria.
2.5.2 Thermal profile for the amplification of PCR
Table 3. The thermal profile of bcfC gene specific primer of Salmonella spp.
Table 4. The thermal profile for invC gene specific primer Shigella spp.
Table 5. Thermal profile for 16S rRNA gene specific primer of E. coli
Table 6. Thermal profile for groEL gene specific primer of Vibrio spp.
2.6 Antibiotic sensitivity testing
The antimicrobial susceptibility assay was detected using the disc diffusion method in accordance with the Clinical and Laboratory Standards Institute's (CLSI, 2016 ![]() ) guidance. The disc diffusion method was used to test antimicrobial drug susceptibility against 12 widely used antibiotics (Bauer et al., 1966
) guidance. The disc diffusion method was used to test antimicrobial drug susceptibility against 12 widely used antibiotics (Bauer et al., 1966 ![]() ). The following antibiotic discs were employed for Salmonella spp., Shigella spp., E. coli, and Vibrio spp.: Amoxycillin (AMX, 20 μg), Gentamicin (GEN, 10 μg), Amikacin (AK, 30 μg), Ceftriaxone (CTR, 30 μg), Ceftazidime (CAZ, 30 μg), Azithromycin (AZM, 15 μg), Levofloxacin (LE, 5 μg), Colistin (CL, 10 μg), Cefepime (CPM, 30 μg), Aztreonam (AT, 30 μg), Ampicillin (AMP, 10 μg), Erythromycin (E, 15 μg) Doxycycline (DO, 30 μg), Cefixime (CPM, 30 μg).
). The following antibiotic discs were employed for Salmonella spp., Shigella spp., E. coli, and Vibrio spp.: Amoxycillin (AMX, 20 μg), Gentamicin (GEN, 10 μg), Amikacin (AK, 30 μg), Ceftriaxone (CTR, 30 μg), Ceftazidime (CAZ, 30 μg), Azithromycin (AZM, 15 μg), Levofloxacin (LE, 5 μg), Colistin (CL, 10 μg), Cefepime (CPM, 30 μg), Aztreonam (AT, 30 μg), Ampicillin (AMP, 10 μg), Erythromycin (E, 15 μg) Doxycycline (DO, 30 μg), Cefixime (CPM, 30 μg).
3. Results and Discussion
3.1 Cultural, staining, and biochemical characteristics
3.1.1 Bacterial growth on nutrient agar
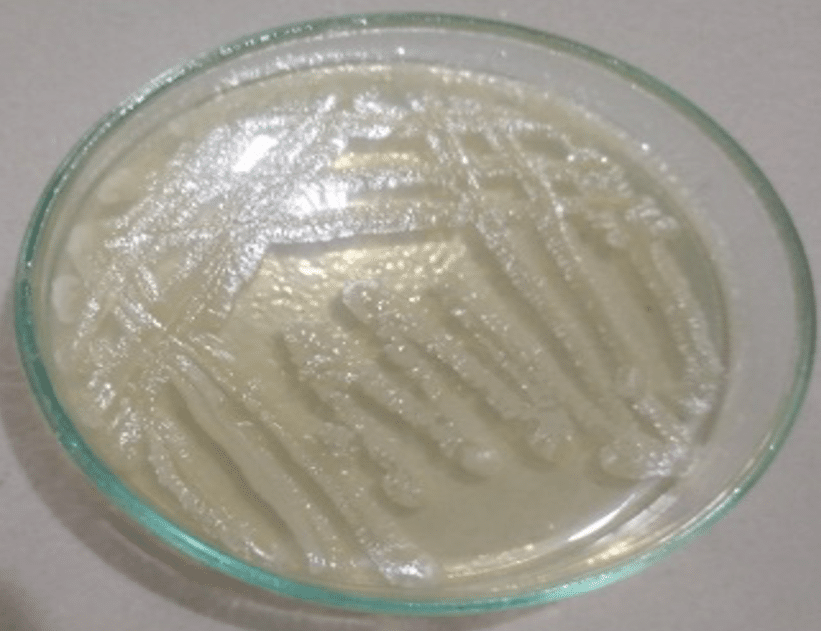
The appearance of homogenous turbidity in the nutrient broth indicated the growth of bacteria. The appearance of various types of colony characteristics (large, circular, convex, greyish-white, smooth, and translucent) on nutrient agar media indicated the presence of mixed bacterial population. Then each type of colony was sub-cultured on different selective media for specific bacterial colony as well as to obtain pure culture of Salmonella spp., Shigella spp., E. coli, and Vibrio spp. (Figure 2).
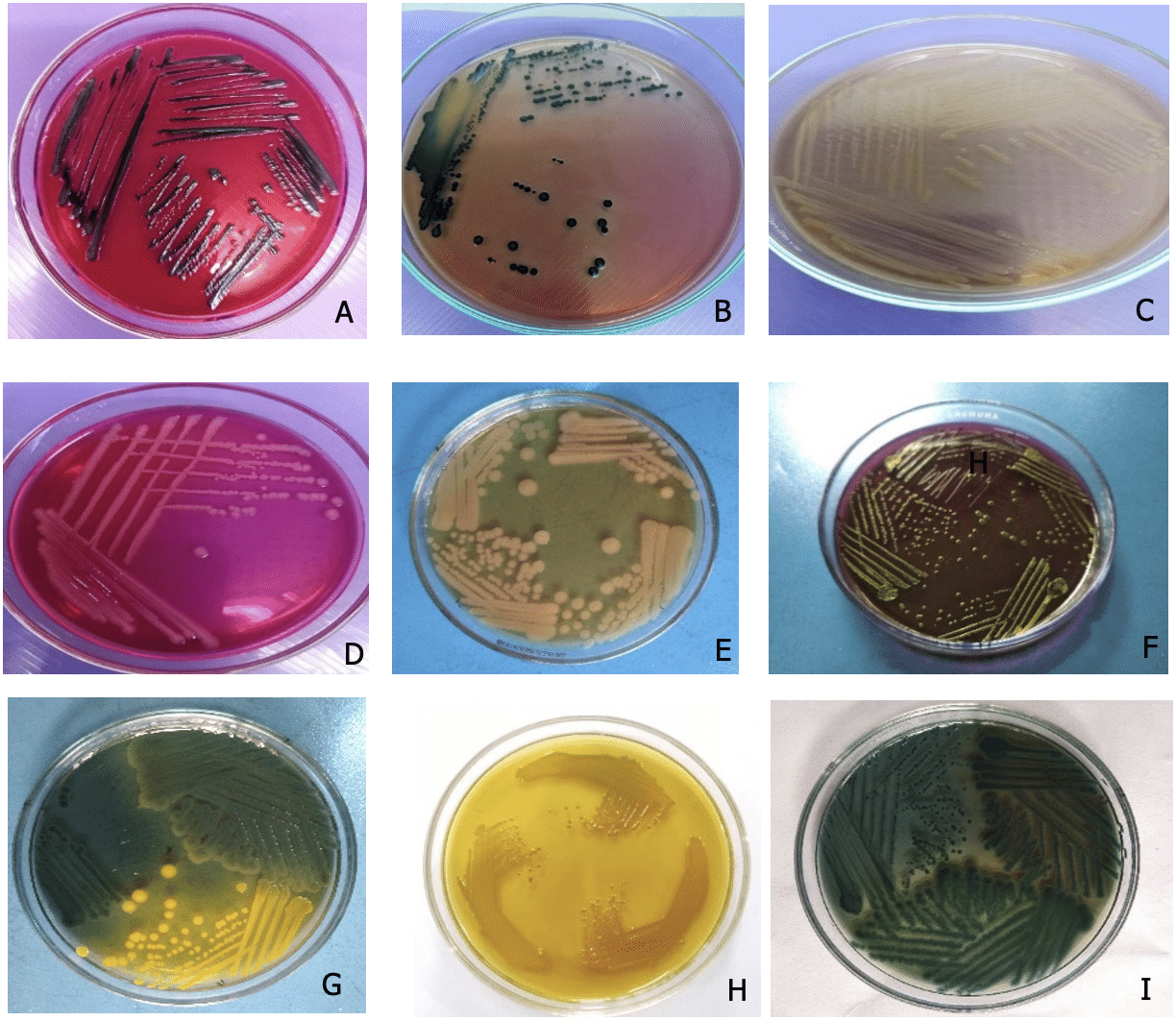
3.1.2 Subculture of bacterial isolates on different media for obtaining pure isolates of Salmonella spp., Shigella spp., E. coli, and Vibrio spp.
Among the 40 samples, 3 isolates showed colony characteristics consistent with Salmonella spp. on various media such as SS, XLD, BGA, and MAC Agar. On Salmonella Shigella (SS) agar media, all six suspected Shigella isolates formed characteristic big, round, convex, and colorless colonies. All 14 probable E. coli isolates formed smooth, round, black or green colonies with a metallic sheen on EMB agar. Following primary culture, all probable Vibriospp. isolates (n=3) formed characteristic smooth, round, yellow or greenish colonies on TCBS media.
Under the microscope, all of the putative Salmonella, Shigella, E. coli, and Vibrio isolates appeared to be gram-negative, pink-colored, tiny rod-shaped bacteria organized singly or in pairs. Salmonella isolates that are culture positive and urease positive change color from yellow to purple-red extremely quickly after organisms are inoculated into test tubes. However, unfavorable reactions that occur slowly imply positive results for Salmonella spp. All of the suspected Salmonella isolates tested negative for urease. The positive Shigella isolates did not ferment dextrose, maltose, lactose, or sucrose in comparison to gas generation. The isolates were only mannitol-positive, indicating acid generation.
The suspected Shigella isolates tested positive for Methyl Red and the indole test. The suspected Shigella isolates produced a pink ring and tested negative for VP.
The positive isolates of E. coli and Vibrio spp. fermented dextrose, maltose, lactose, and sucrose, as well as produced acid and gas. All probable E. coliisolates were positive for the Methyl Red test. However, Vibrio spp. isolates were negative for the Methyl Red test. Both putative isolates were VP-positive (Figure 3).
3.2 Molecular confirmation of suspected Salmonella spp., Shigella spp., E. coli, and Vibrio spp. isolates
DNA from all probable Salmonella isolates was used in the PCR assay. DNA from pure culture was extracted using the standard boiling procedure.Salmonella spp. were validated by amplification of the bcfC gene (993 bp). Finally, 3 isolates were identified as salmonella spp. (Figure 4 A). Among the 40 samples, 6 were positive for Shigella spp. using a genus-specific primer of the invC gene. The amplicon size of 875 bp was observed using the UV trans-illuminator (Figure 4 B). Of the 40 samples analyzed, 14 tested positive for E. coli using a primer specific to the 16S rRNA gene. The resulting amplicon, measuring 704 bp, was detected with a UV trans-illuminator (Figure 4C).
Out of the 10 isolates initially suspected to be Vibrio, 3 were confirmed as Vibrio species using genus-specific groEL primers, which yielded a positive band at 1117 bp (Figure 4D). Further analysis with multiplex PCR and species-specific groEL primers identified all three confirmed isolates as Vibrio cholerae, producing a positive band at 418 bp (Figure 4E). No isolates were identified as Vibrio parahaemolyticus, Vibrio alginolyticus, or Vibrio vulnificusthrough multiplex PCR.
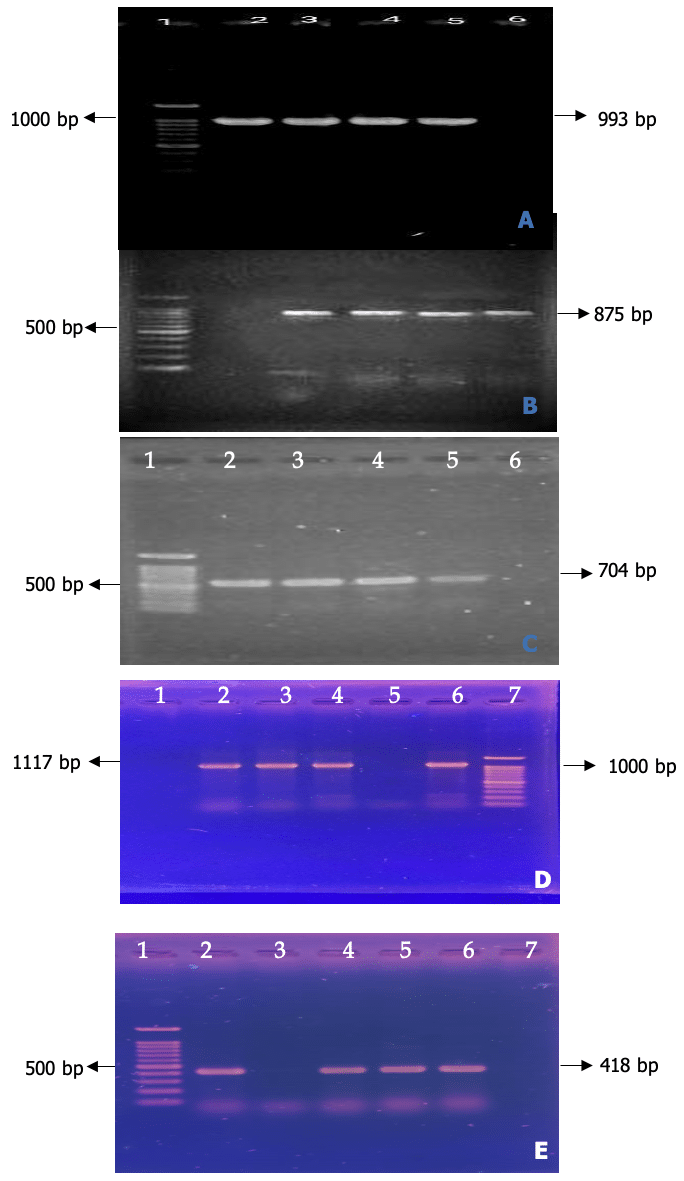 Figure 4 A. Amplification of bcfC gene for Salmonella spp. (993bp), Lane 1: 100bp DNA ladder, Lane 2: Positive control, 3-5: Salmonella spp. Figure 4 B. PCR amplification of Shigella genus specific gene primer (875 bp) of Shigella spp. Lane 1: 100bp DNA Marker, Lane 1-4: Representative Shigella spp. isolates. Figure 4 C. PCR amplification of E. coli genus specific gene primer (16S rRNA) of E. coli. Lane 1: 100bp DNA Marker, Lane 2-4: Representative E. coli isolates. Figure 4 D. PCR amplification of groEL gene for specific detection of the genus Vibrio Lane 1: 100bp DNA Marker, Lane-2: Positive control. Lane-3: Negative control, Lane 4-6: Representative Vibrio isolates. Figure 4 E. Amplification of groEL gene for the specific detection of V. alginohyticus (301), V. parahaemolyticus (644), V. cholerae (418) and V. vulnificus (192).
Figure 4 A. Amplification of bcfC gene for Salmonella spp. (993bp), Lane 1: 100bp DNA ladder, Lane 2: Positive control, 3-5: Salmonella spp. Figure 4 B. PCR amplification of Shigella genus specific gene primer (875 bp) of Shigella spp. Lane 1: 100bp DNA Marker, Lane 1-4: Representative Shigella spp. isolates. Figure 4 C. PCR amplification of E. coli genus specific gene primer (16S rRNA) of E. coli. Lane 1: 100bp DNA Marker, Lane 2-4: Representative E. coli isolates. Figure 4 D. PCR amplification of groEL gene for specific detection of the genus Vibrio Lane 1: 100bp DNA Marker, Lane-2: Positive control. Lane-3: Negative control, Lane 4-6: Representative Vibrio isolates. Figure 4 E. Amplification of groEL gene for the specific detection of V. alginohyticus (301), V. parahaemolyticus (644), V. cholerae (418) and V. vulnificus (192). 3.3 Antibiotics sensitivity test of isolated Salmonella spp. Shigella spp., E. coli, and Vibrio spp.
Among the three Salmonella spp. isolates, all were resistant to Amoxicillin, 70% to Ceftazidime, 33% to Ceftriaxone, and 25% to Levofloxacin. They were sensitive or intermediately sensitive to Azithromycin, Colistin, Amikacin, and Gentamicin.
For the six Shigella spp. isolates, 83% were resistant to Cefepime and 67% to Ceftazidime. They showed high or intermediate sensitivity to Azithromycin, Amoxicillin, Ceftriaxone, Colistin, Amikacin, and Gentamicin.
All 14 E. coli isolates displayed resistance patterns with 100% resistance to Amoxicillin, 93% to Colistin, 72% to Azithromycin, Cotrimoxazole, and Cefepime, and 57% to Gentamicin. They reacted to ceftriaxone and amikacin with high or intermediate sensitivity. Among the three Vibrio spp. isolates, 100% were resistant to Ampicillin, 81% to Cefepime, 72% to Cefixime, 70% to Amoxicillin, and 67% to both Erythromycin and Ceftazidime. They had high or intermediate sensitivity to amikacin, gentamicin, levofloxacin, doxycycline, and cotrimoxazole.
Table 7. The percentages of antibiotics sensitivity and resistance patterns of Salmonella spp. (n=3) isolates against different antibiotic classes.
Table 8. The percentages of antibiotics sensitivity and resistance patterns of Shigella spp. (n=6) isolates against different antibiotic classes.
Table 9. The percentages of antibiotics sensitivity and resistance patterns of E. coli (n=14) isolates against different antibiotic classes.
Table 10. The percentages of antibiotics sensitivity and resistance patterns of Vibrio cholerae (n=3) isolates against different antibiotic classes.
4. Discussion
In the fields of industry, research, and medicine, the Enterobacteriaceae family of bacteria is one of the most significant (Ramírez-Castillo et al., 2015). Microorganisms play a significant role in maintaining the quality of water and have the potential to spread pathogenic bacteria, viruses, and parasites into drinking water (Cabral, 2010; Tebbutt, 1977). Microorganisms like Salmonella, Shigella, Escherichia coli, and Vibrio cholerae are among those that can make people sick when they consume polluted water (Cabral, 2010).
This study intended to examine the presence, cultural characteristics, biochemical test, molecular characteristics, and antibiotic susceptibility status of Salmonella spp., Shigella spp., E. coli, and Vibrio spp. strains isolated from different water sources of Mymensingh city in Bangladesh.
Table 11. The overall occurrence of Salmonella spp., Shigella spp., E. coli, and Vibrio spp. in river water, pond water, tap water, and tube-well water.
The Enterobacteriaceae family is crucial in medicine, industry, and research (Ramírez-Castillo et al., 2015 ![]() ). Microorganisms in water can transfer diseases, including bacteria, viruses, and parasites (Cabral, 2010
). Microorganisms in water can transfer diseases, including bacteria, viruses, and parasites (Cabral, 2010 ![]() ; Tebbutt, 1977
; Tebbutt, 1977 ![]() ). Pathogens like Salmonella, Shigella, E. coli, and Vibrio cholerae can contaminate drinking water (Cabral, 2010
). Pathogens like Salmonella, Shigella, E. coli, and Vibrio cholerae can contaminate drinking water (Cabral, 2010 ![]() ).
).
This study focused on assessing the presence, characteristics, and antibiotic resistance of Salmonella, Shigella, E. coli, and Vibrio strains from different water sources in Mymensingh city, Bangladesh.
The occurrence of 7.5% of Salmonella spp. in water was comparatively lower than the previously conducted reports of 19.4% and 3.3% of S. enterica in pond and well water, respectively, by Gu et al. (2021 ![]() ), 0.89% by Momtaz et al. (2013
), 0.89% by Momtaz et al. (2013 ![]() ), and 4% by Dekker et al. (2018
), and 4% by Dekker et al. (2018 ![]() ). Similarly, the higher findings were reported in southern Georgia and northern Florida at 28.2% by Luo et al. (2015
). Similarly, the higher findings were reported in southern Georgia and northern Florida at 28.2% by Luo et al. (2015 ![]() ) and on the banks of the Zarqa River at 12.8% by Tarazi et al. (2021
) and on the banks of the Zarqa River at 12.8% by Tarazi et al. (2021 ![]() ). These fluctuations could be brought on by changes in the seasonal and geographic conditions along the Zarqa River's banks. A total of 3 isolates from the water samples utilized in this investigation were recognized as Salmonella spp. based on cultural and biochemical traits. According to Mridha et al. (2020
). These fluctuations could be brought on by changes in the seasonal and geographic conditions along the Zarqa River's banks. A total of 3 isolates from the water samples utilized in this investigation were recognized as Salmonella spp. based on cultural and biochemical traits. According to Mridha et al. (2020 ![]() ), the colony features of Salmonella spp. that were isolated in various media resembled those of Salmonella spp. Variations in the outcomes could be caused by genetic variables and the type of organisms residing there. The isolates of Salmonella spp. exhibited higher resistance patterns against Amoxicillin (100%), Ceftazidime (70%), and Cephalexin (67%). Furthermore, a lower level of resistance was noted against Levofloxacin (25%), Ceftriaxone (33%) and almost no resistance or highly susceptibility to Cefepime, Amikacin, Gentamicin, Azithromycin, Colistin. The previous studies reported the higher resistance patterns of Salmonella spp. against amoxicillin by Mian et al. (2020
), the colony features of Salmonella spp. that were isolated in various media resembled those of Salmonella spp. Variations in the outcomes could be caused by genetic variables and the type of organisms residing there. The isolates of Salmonella spp. exhibited higher resistance patterns against Amoxicillin (100%), Ceftazidime (70%), and Cephalexin (67%). Furthermore, a lower level of resistance was noted against Levofloxacin (25%), Ceftriaxone (33%) and almost no resistance or highly susceptibility to Cefepime, Amikacin, Gentamicin, Azithromycin, Colistin. The previous studies reported the higher resistance patterns of Salmonella spp. against amoxicillin by Mian et al. (2020 ![]() ). These differences may be due to geographical and seasonal variations.
). These differences may be due to geographical and seasonal variations.
The occurrence of 15% of Shigella spp. in water samples was higher than the previously conducted report of 5.4% by Hsu et al. (2010 ![]() ). Similarly, the higher 27% by Saima et al. (2018
). Similarly, the higher 27% by Saima et al. (2018 ![]() ). These discrepancies may arise from differences in sample numbers, methods employed throughout the process, and regional and seasonal distribution. Using drinking water samples from this study, 6 isolates were determined to be Shigella spp. based on cultural and biochemical traits. According to Makabanyane et al. (2015
). These discrepancies may arise from differences in sample numbers, methods employed throughout the process, and regional and seasonal distribution. Using drinking water samples from this study, 6 isolates were determined to be Shigella spp. based on cultural and biochemical traits. According to Makabanyane et al. (2015 ![]() ), the colony features of Shigella spp. isolated in various media mirror those of Shigella spp. The Shigella spp. isolates showed higher resistance patterns to ceftazidime (67%) and cefepime (83%). Furthermore, a lower level of resistance was noted against Amoxicillin (17%), Colistin (10%) and almost no resistance or highly susceptibility was show to Azithromycin, Levofloxacin, Cotrimoxazole, Ceftriaxone, Amikacin, and Gentamicin. According to Matloko et al. (2021
), the colony features of Shigella spp. isolated in various media mirror those of Shigella spp. The Shigella spp. isolates showed higher resistance patterns to ceftazidime (67%) and cefepime (83%). Furthermore, a lower level of resistance was noted against Amoxicillin (17%), Colistin (10%) and almost no resistance or highly susceptibility was show to Azithromycin, Levofloxacin, Cotrimoxazole, Ceftriaxone, Amikacin, and Gentamicin. According to Matloko et al. (2021 ![]() ), who reported that Shigella spp. had greater resistance patterns to cefuroxime (36.1%, 13/36), cefepime (30.6%, 11/36), cefuroxime, and aztreonam (27.8%, 10/36) than to the other antibiotics examined (susceptibilities of > 75%), etc.
), who reported that Shigella spp. had greater resistance patterns to cefuroxime (36.1%, 13/36), cefepime (30.6%, 11/36), cefuroxime, and aztreonam (27.8%, 10/36) than to the other antibiotics examined (susceptibilities of > 75%), etc.
The occurrence of 35% of E. coli in water samples was higher than previously conducted reports of 7.5 8% by Momtaz et al. (2013 ![]() ). Similarly, the higher findings found 60.3% by Thani et al. (2016
). Similarly, the higher findings found 60.3% by Thani et al. (2016 ![]() ) and 77.97% from Larut River were commensal strains, with phylogroup B1 (39.55%) and phylogroup A (38.42%) by Bong et al. (2022
) and 77.97% from Larut River were commensal strains, with phylogroup B1 (39.55%) and phylogroup A (38.42%) by Bong et al. (2022 ![]() ). Variations in sample numbers, methods types employed throughout the process, and regional and seasonal dispersion could all contribute to these discrepancies. Utilizing drinking water samples for this investigation, a total of 16 isolates were determined to be E. colibased on biochemical and cultural traits. According to Schaumburg et al . (2015
). Variations in sample numbers, methods types employed throughout the process, and regional and seasonal dispersion could all contribute to these discrepancies. Utilizing drinking water samples for this investigation, a total of 16 isolates were determined to be E. colibased on biochemical and cultural traits. According to Schaumburg et al . (2015 ![]() ) and Hossain et al. (2020
) and Hossain et al. (2020 ![]() ), the colony properties of the isolated E. coli in various media reflect those of E. coli. The higher resistance patterns were shown by the E. coli isolates to Amoxicillin (100%), Colistin (93%), Azithromycin (72%), Cotrimoxazole (72%), Cefepime (72%), Ceftazidime (57%), and Gentamicin (57%). Furthermore, a lower level of resistance was noted against Levofloxacin (7%), Ceftriaxone (36%), Amikacin (29%). Prior research has documented the 49.21% of the Escherichia coli isolates were susceptible, 12.90% were intermediate, and 37.90% were resistant overall. Vancomycin (94.64%) and erythromycin (85.71%) both exhibited high resistance. There was also a significant level of susceptibility to ceftrioxine (89.29%), gentamicin (91.07%), and ciprofloxacin (94.64%). Adzitey et al. (2016) who reported that a comparatively greater proportion of the Escherichia coli isolates from the water sample (50%) showed intermediate resistance to amoxycillin/clavulanic acid. Variations may be due to differences in geography, seasonality, sample sizes, and methods.
), the colony properties of the isolated E. coli in various media reflect those of E. coli. The higher resistance patterns were shown by the E. coli isolates to Amoxicillin (100%), Colistin (93%), Azithromycin (72%), Cotrimoxazole (72%), Cefepime (72%), Ceftazidime (57%), and Gentamicin (57%). Furthermore, a lower level of resistance was noted against Levofloxacin (7%), Ceftriaxone (36%), Amikacin (29%). Prior research has documented the 49.21% of the Escherichia coli isolates were susceptible, 12.90% were intermediate, and 37.90% were resistant overall. Vancomycin (94.64%) and erythromycin (85.71%) both exhibited high resistance. There was also a significant level of susceptibility to ceftrioxine (89.29%), gentamicin (91.07%), and ciprofloxacin (94.64%). Adzitey et al. (2016) who reported that a comparatively greater proportion of the Escherichia coli isolates from the water sample (50%) showed intermediate resistance to amoxycillin/clavulanic acid. Variations may be due to differences in geography, seasonality, sample sizes, and methods.
The occurrence of 7.5% of Vibrio spp. in water samples was higher than previously conducted reports of 1.7% by Alam et al. (2014 ![]() ). Similarly, higher findings found 89% by Shanan et al. (2011
). Similarly, higher findings found 89% by Shanan et al. (2011 ![]() ), 35% by Ferdous et al. (2018
), 35% by Ferdous et al. (2018 ![]() ). These differences could be the result of changes in sample numbers, methods utilized throughout the entire process, and regional and seasonal distribution. There were 3 distinct Vibrio spp. isolates found according to the cultural and biological traits of the drinking water samples that were utilized for this investigation. Another study conducted by Azwai et al. (2016
). These differences could be the result of changes in sample numbers, methods utilized throughout the entire process, and regional and seasonal distribution. There were 3 distinct Vibrio spp. isolates found according to the cultural and biological traits of the drinking water samples that were utilized for this investigation. Another study conducted by Azwai et al. (2016 ![]() ) reported that the colony properties of isolated Vibrio spp. in various media are similar to those of Vibrio spp., and the higher resistance patterns against 100% Ampicillin, 81% Cefepime, 72% Cefixime, 70% Amoxicillin, 67% Erythromycin, and 67% Ceftazidime were shown by Vibrio spp. Conversely, the higher sensitive against Gentamicin, Levofloxacin, Doxyciline, Cotrimoxazole, Amikacin.
) reported that the colony properties of isolated Vibrio spp. in various media are similar to those of Vibrio spp., and the higher resistance patterns against 100% Ampicillin, 81% Cefepime, 72% Cefixime, 70% Amoxicillin, 67% Erythromycin, and 67% Ceftazidime were shown by Vibrio spp. Conversely, the higher sensitive against Gentamicin, Levofloxacin, Doxyciline, Cotrimoxazole, Amikacin.
5. Conclusions
The present study was carried out to isolate and identify Salmonella spp., Shigella spp., E. coli and Vibrio spp. from water samples of Mymensingh city. In addition, their antibiotic resistance profile was also determined at the phenotypic and genotypic level. The water sources from various locations in Mymensingh municipality are contaminated with enteric bacteria, posing a significant public health risk to residents who rely on the water for drinking. The presence of bacterial species such as Salmonella, Shigella, E. coli, and Vibrio highlights the potential for gastrointestinal diseases. The antibiotic resistance profiles of these isolates raise further concern. High levels of resistance to commonly used antibiotics like amoxicillin and ceftazidime suggest that treatment of infections caused by these bacteria may be challenging, and the continued use of antibiotics could exacerbate antimicrobial resistance. However, the isolates showed varied sensitivity to other antibiotics, indicating potential therapeutic options for managing infections. In conclusion, the study underscores the need for urgent measures to improve water quality, implement proper sanitation practices, and develop antimicrobial stewardship programs to address both contamination and the growing issue of antibiotic resistance in the region.
Acknowledgements
The authors would like to thank the Ministry of Science and Technology, Government of the People’s Republic of Bangladesh (Ref. No. 39.00.0000.009.14.019.21-745, 417-ES) for supporting by giving financial funds to facilitate the research work.
Data availability statement
The data generated from this study will be available on the valid request from the corresponding author.
Informed consent statement
Not applicable.
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
Conceptualization: Mohammad Ferdousur Rahman Khan; Data collection, study conduct, and manuscript write up: Dula chakraborty and Limon Biswas; Methodology preparation: Mahbubul Pratik Siddique and Najmun Nahar Popy; Formal analysis, data analysis, first draft develop, and result section interpretation: Mohammad Ferdousur Rahman Khan, Mahbubul Pratik Siddique, Dula chakraborty, and Shantono Acharjee; Figure preparation, formal analysis, reviewed, and revised the final version of the manuscript: Dula chakraborty, Limon Biswas, Shantono Acharjee, and Najmun Nahar Popy, and Mohammad Ferdousur Rahman Khan. All authors critically reviewed the manuscript and agreed to submit final version of the article.



