Over recent decades, the consumption of fast food has significantly risen in most urban areas of the world and, accordingly, in Bangladesh too, because fast foods are time-saving and cheap, and lifestyles are changing. However, fast foods remain a major public health concern owing to bacterial contamination, especially by pathogenic bacteria like E. coli and S. aureus (Hossain et al., 2024; Ahmed et al., 2014; Bondi et al., 2014). E. coli is a Gram-negative bacterium located in the intestines of both humans and animals. There have been instances in Bangladesh where foodborne outbreaks, including those associated with E. coli and other enteric pathogens, have been linked to fast food. The outbreaks are often associated with contaminated meat, particularly beef and poultry, which are common ingredients in fast food items such as burgers, sandwiches, and fried chicken. The majority of E. coli strains were avirulent; however, some strains are pathogenic, like the ones producing Shiga toxins that cause severe gastrointestinal diseases (Rana et al., 2024; Bruyand et al., 2018). Additionally, S. aureus is a gram-positive bacterium that is responsible for a number of different infections; it causes foodborne illness too due to heat-stable enterotoxins production (Julqarnain et al., 2022). These could contaminate food during preparation, handling, or storage in unhygienic conditions, which is quite common in fast-food places, especially in developing countries (Ahmed et al., 2014; Ayamah et al., 2021). Staphylococcus spp. is a prevalent cause of foodborne illnesses worldwide (Dehkordi et al., 2019). Furthermore, according to Abebe et al. (2010), the primary cause of staphylococcal food poisoning (SFP) is the intake of foods infected with S. aureus. Additionally, it is harmful due to its zoonotic characteristics (Rahman et al., 2020). Symptoms of food poisoning (SFP) is typically associated with poultry and its products, meat and meat products, bakery items (such as pizza, burgers, pastries, and buns), as well as salads (Argudín et al., 2010). This pathogen's presence poses a significant risk to consumers and results in economic losses due to the spread of diseases that are transmitted through food. It is not uncommon for those who have SFP to experience symptoms like nausea, stomach cramps, vomiting, and even diarrhea (Bondi et al., 2014). According to Tong et al. (2015), the majority of staphylococcal infections are caused by S. aureus, which is a leading pathogen responsible for food poisoning and skin infections, as well as pneumonia, endocarditis, and toxic shock syndrome in humans. Likewise, research has shown that ready-to-eat (RTE) foods can be contaminated with E. coli (Abebe et al., 2020). In a previously study found that this species can live on various surfaces, including hands, and is easily spread to foods (Ayamah et al., 2021). E. coli serves as a key indicator of contamination by fecal and enteric pathogens. While most E. coli strains are harmless, certain types are recognized for their ability to lead to serious gastrointestinal issues in humans, including HC and HUS. Shiga toxins (stx1 and stx2), enterohemolysin (hlyA), and intimin (eaeA) have been identified as virulence factors that play a significant role in the pathogenesis of these disorders (Bruyand et al., 2018). Additional studies have been conducted on E. coli and other pathogens in RTE foods (Secim and Ucar, 2018). Moreover, within the last couple of years, there has been an alarming rise in antibiotic-resistant bacteria in some of the most common human pathogens, including E. coli and S. aureus. The general misuse and overuse of antibiotics in either clinical or agricultural settings have accelerated MDR strain development, infections of which are becoming progressively difficult to treat (Mow et al., 2024; Founou et al., 2016). In Bangladesh, MDR bacteria are pointed out in food products; this calls for improvement in the safety of foods and AM stewardship (Noor and Feroz, 2016). In above statement there was no data about virulence gene detection that causes foodborne illness of human being. Moreover, in Bangladesh, fast food is the best food choice for all age’s peoples. From the above statement, it is clear that pathogenic E. coli and S. aureus have great public health hazards and infect human health. The point of this study was to find out how common E. coli and S. aureus are in fast food in Gazipur City, Bangladesh, as well as how resistant they are to a number of common antibiotics and how to find their virulence genes using PCR to target specific genes. To the best of our understanding and consciousness. This study will add to the knowledge about food safety risks and the increasing problem of antibiotic resistance in Bangladesh.
2. Materials and Methods
2.1 Ethical approval
No ethical approval is required for this study.
2.2 Study area and sample collection
The current research was designed for January to June 2023 and conducted in the Department of Microbiology Laboratory of Gono Bishwabidyalay, Savar, Dhaka, Bangladesh. For this research, different selected areas (Chourasta more, Tongi College Gate, Shaheed Tazuddin Medical College Hospital, Passport Office) in Gazipur city, Bangladesh, were selected. A total of 120 fast food samples, including burgers, sandwiches, chicken rolls, and fuchka were collected from different street vendors and restaurants in aseptic conditions using sterile bags (transparent sealed zip-lock polybags; thickness 30-100 mic, size 175 × 100 mm) and then samples were transported to the Department of Microbiology, Gono Bishwabidyalay, Savar, Dhaka-1344, Bangladesh under chilled conditions (4 °C) for immediate processing and bacteriological analysis. The map of the study area was created by Acrgis software.
2.3 Sample preparation
Solid samples of fuchka, burger, sandwich and chicken roll coat and its contents were blended using sterile morter and pestle followed by dissolving with distilled water at 1:10 ratio. Then dissolved samples were centrifuged at 1500 rotate per minute (rpm) (Biobase, China). The supernatant was collected for culturing of S. aureus and E. coli on selective culture media.
 Figure 1. Map of the study area (Gazipur).
Figure 1. Map of the study area (Gazipur).
2.4 Isolation and identification of bacteria
For the purpose of isolating bacteria, 10 grams of each food sample were homogenized in 90 milliliters of sterile phosphate-buffered saline (PBS) and then exposed to serial dilution (PBS; pH: 7.4, Merck KGaA, Germany). The diluted samples were subsequently plated on MacConkey agar (Hi Media Private Ltd., India) and EMB agar (Hi Media Private Ltd., India) for the selective isolation of E. coli and on Mannitol Salt Agar (Hi Media Private Ltd., India) for the isolation of S. aureus. These plates were incubated at 37 °C for 24 to 48 hours. Colonies displaying typical morphology were selected for identification (Cheesbrough, 2006). Biochemical tests of E. coli isolates included the indole test, methyl red test, Voges-Proskauer test, and citrate utilization test, which confirmed the identification. On the other hand, catalase and coagulase tests were conducted for S. aureus confirmation. Confirmation at the molecular level was also done by performing PCR targeting the malB gene in E. coli and the nuc gene of S. aureus (Queipo-Ortuño et al., 2008; Aklilu et al., 2016).
2.5 Molecular confirmation of E. coli and S. aureus
2.5.1 Extraction of genomic DNA (S. aureus and E. coli) by boiling method and thermal profiling
Following the instructions provided by Queipo-Ortuño et al. (2008), the DNA template was produced using the boiling process. A solitary colony of S. aureus was extracted from 100 microliters of distilled water and placed in an eppendorf tube. The mixture was thoroughly mixed and then subjected to boiling for a duration of 10 minutes. Following the boiling process, the tubes were immediately cold-shocked for 10 minutes at a temperature of 4 °C, and the sample was subsequently subjected to centrifugation at a velocity of 10,000 rpm for a duration of 10 minutes. The supernatant was collected, and it was subsequently utilized as a template for DNA. Similarly, DNA of E. coli was isolated from selected colonies by the boiling method (Queipo-Ortuño et al., 2008). The malB gene (585 bp) and nuc gene (279 bp) were amplified by PCR to confirm E. coli and S. aureus respectively. A total of 25 µL reaction mixture including 2 µL of DNA temple, 12.5 µL of 2X master mix, (Go taq green master mix, Promega, Dane Country, WI, USA), 0.5 µL of each forward and reverse primer, 9.5 µL nuclease free water was adjusted as PCR assays. The amplified products were further analyzed for the expected band sizes using 1.5% (w/v) agarose gel electrophoresis (Aklilu et al., 2016). The primer details of S. aureus and E. coli are mentioned in Table 1, and for E. coli in Table 2. A 100 base pair DNA ladder (Thermo Fisher, USA) was used as a size marker.
2.6 Antibiotic Sensitivity Testing (AST)
According to CLSI (2021) recommendations, antibiotic susceptibility testing via the Kirby-Bauer disc diffusion method was applied. 13 commercially available antibiotics, including Azithromycin (15 µg), Penicillin (10 µg), Oxacillin (1 µg), Cefoxitin (30 µg), Ciprofloxacin (5 µg), Norfloxacin (10 µg), Levofloxacin (5 µg), Doxycycline (30 µg), Erythromycin (5 µg), Gentamycin (5 µg), Clindamycin (2 µg), Tetracycline (30 µg), and Vancomycin (30 µg) were applied for S. aureus. Similarly, commercially available 10 antibiotics, including ampicillin (25 µg), colistin (10 µg), ciprofloxacin (5 µg), gentamicin (10 µg), streptomycin (10 µg), ceftazimide (30µg), tetracycline (30 µg), chloramphenicol (10 µg), and meropenem (10 µg), were tested against E. coli. The bacterial inoculum was prepared and adjusted to a 0.5 McFarland turbidity standard, then spread on Mueller-Hinton agar (Hi Media Private Ltd., India) and incubated overnight at 37 °C (Biobase, China). After 24 hours, the zone of inhibition was measured and interpreted with a millimeter scale by CLSI standards (CLSI, 2021). In current research, more than three antibiotics showed resistance against E. coli and S. aureus indicated Multi-drug Resistance (MDR) followed by previously published studies (Titilawo et al., 2015; Adzitey et al., 2012).
Table 1. Oligonucleotide primer sequences for detection of S. aureus and E. coli.
Table 2. Thermal profile for the amplification of nuc gene of S. aureus and malB gene of E. coli.
2.7 Statistical analysis
We used to excel version 24.0, to analyze the data. Descriptive statistics were used to describe the prevalence of bacterial contamination and antibiotic resistance. Prevalence, virulence gene, distribution of E. coli and S. aureus were detected by data analysis.
3. Results
3.1 Prevalence of S. aureus and E. coli in fast food samples
A total of 120 fast food samples (fuchka, burger, sandwich, and chicken roll) were collected from various 10 types of restaurants and shops in Gazipur City. Out of 120 samples, 26 (21.66%) were positive for S. aureus, whereas E. coli was confirmed in 22 (18.33%) samples, respectively. The highest prevalence rate of S. aureus was recorded in fuchka 12 (40%), followed by sandwich 6 (20%) and Burger and Chicken Roll 4 (13.33%), respectively (Table 3). On the other hand, the highest number of positive cases of E. coli were found in fuchka 10 (33.33%), while the lowest prevalence was detected in burger and chicken roll 2 (6.66%), respectively. Out of 48 positive isolates, 26 S. aureus phenotypically identified by cultural and biochemical tests whereas 12(46.15%) were genotypically detected. On the other hand, E. coli was detected genotypically 10 (45.45%) whereas 22 isolates were found phenotypically respectively (Figure 2). Additionally, in our study the highest number of S. aureus was recorded in fuchka samples from Chourasta more, Gazipur whereas highest number of E. coli was found in same location in fuchka samples. On the other hand, the lowest prevalence of S. aureus in Chicken roll and Burger was recorded in Tongi College Gate and Passport office, Gazipur respectively (Table 4).
3.2 Phenotypic characteristics of S. aureus and E. coli
After primary culture on nutrient agar and nutrient broth, S. aureus was cultured on MSA and observed with yellow colonies on 24 hours of incubation (Figure 3: A). On the other hand, E. coli showed metallic green colonies on selected EMB agar (Figure 3: B). After cultural confirmation, both S. aureus and E. coli go for biochemical tests for further confirmation. S. aureus gave positive reactions for catalase, coagulase, and MR-VP tests and only negative results for the indole test, whereas E. coli demonstrated positive results for the MR test, indole test, sugar fermentation tests (dextrose, maltose, mannitol, sucrose tests), and the VP test negative.
Table 3. Prevalence of S. aureus and E. coli based on different fast food.
Table 4. Occurrence of E. coli and Staphylococcus spp. based on source of location.
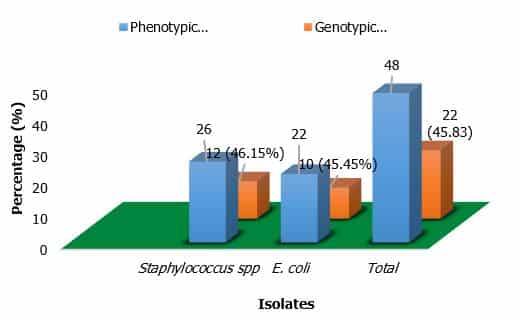 Figure 2. Comparison between phenotypic and genotypic identification
Figure 2. Comparison between phenotypic and genotypic identification
 Figure 3 (A). Yellow colonies of S. aureus in MSA Agar;
Figure 3 (A). Yellow colonies of S. aureus in MSA Agar;
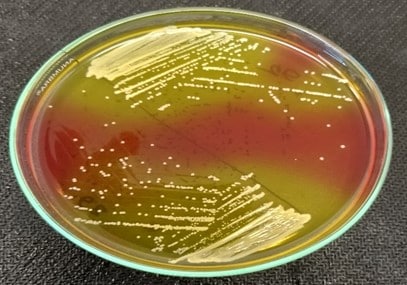 (B). Green metallic sheen colonies of E. coli in EMB Agar.
(B). Green metallic sheen colonies of E. coli in EMB Agar.
3.3 Genotyping detection of S. aureus with specific nuc gene
The phenotypically positive S. aureus was further confirmed by PCR. Out of 26 isolates, 12 (46.15%) isolates were confirmed as S. aureus by amplification of nuc gene amplicon size 279 bp (Figure 4). After the confirmation of S. aureus by PCR, the isolates were tested for antibiotic sensitivity phenotypically by the disc diffusion method.
3.4 Molecular detection of E. coli with specific malB gene
Among the 120 samples, 22 were phenotypically identified and 10 (45.45%) found positive for E. coli by PCR targeting malB gene. The amplicon size of 585 bp was visualized under the ultraviolet trans-illuminator (Figure 5).
 Figure 4. Amplification of 279 bp fragment of nuc gene of S. aureus by PCR. Lane M: 100 bp size DNA marker; lane 1-7: DNA samples of S. aureus extracted from fast food sample isolates; lane PC: positive control; lane NC: negative control.
Figure 4. Amplification of 279 bp fragment of nuc gene of S. aureus by PCR. Lane M: 100 bp size DNA marker; lane 1-7: DNA samples of S. aureus extracted from fast food sample isolates; lane PC: positive control; lane NC: negative control.
 Figure 5. PCR amplification of malB gene of E. coli. Lane 1: 1 kb DNA Marker; Lane 2: Positive control; Lane 3: Negative control; and Lane 4-8: Representative E. coli isolates.
Figure 5. PCR amplification of malB gene of E. coli. Lane 1: 1 kb DNA Marker; Lane 2: Positive control; Lane 3: Negative control; and Lane 4-8: Representative E. coli isolates.
3.5 Correlation between biofilm formation and resistance genes
The correlation between biofilm formation and resistant genes was investigated in this research, and the overall biofilm formation was recorded based on high (dark black color), moderate (light red color), and no biofilm formation (no color) correlated with resistant genes. The highest occurrence of biofilm formation in the nuc gene was found 10(83.33%), whereas moderate biofilm formation was only 2(16.66%), and the non-biofilm formation gene was detected 0% respectively (Figure 6).
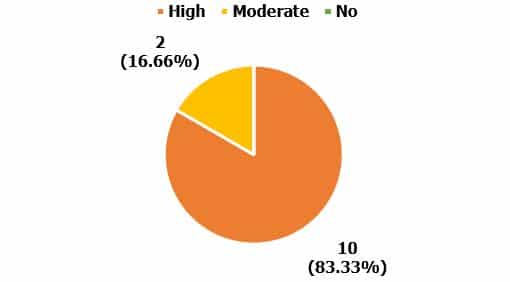 Figure 6. Correlation between biofilm formation and resistance genes (Staphylococcus spp.) High: Orange color; Moderate: Yellow color.
Figure 6. Correlation between biofilm formation and resistance genes (Staphylococcus spp.) High: Orange color; Moderate: Yellow color.
3.6 Antimicrobial susceptibility profile of S. aureus
6 nuc gene positive S. aureus isolates from fast food were tested against 13 different types of antibiotics. All S. aureus strains were resistant to Penicillin, Cefoxitin, Oxacillin 100% and 33.33% of S. aureus isolates were resistant to Vancomycin, Erythromycin, Ciprofloxacin, Tetracycline respectively (Table 6). On the other hand, the highest sensitive elicited against Doxycycline, Gentamycin 100% and Azithromycin, Levofloxacin, tetracycline expressed 83.33% and 66.67%. Vancomycin showed 50% sensitive to all nuc positive isolates of S. aureus respectively (Table 6).
Table 6. Antimicrobial susceptibility of S. aureus isolated from fast food.
3.7 Antimicrobial susceptibility profile of E. coli
All the malB gene-positive E. coli isolates (n=5) were tested against 10 commercially available antibiotics. Only Gentamicin elicited the highest sensitive 60% followed by Chloramphenicol 40% respectively (Table 7). On the other hand, the highest resistance recorded by Erythromycin and Ampicillin (100%) followed by Ceftazidime and Tetracycline (80%) whereas the lowest resistance showed against Colistin, Gentamicin and Imipenem (20%) respectively (Table 7).
Table 7. Antimicrobial susceptibility of E. coli isolated from fast food.
4. Discussion
The significance of food in the spread of various diseases has been well documented over time, particularly in developing nations including Bangladesh, India, Pakistan, Nepal, and Sri Lanka where hygiene standards are often not rigorously adhered to or enforced. The occurrence of these microorganisms can result in numerous foodborne outbreaks. Antimicrobial-resistant bacteria have also spread widely throughout the environment as a result of the widespread use of antibiotics (Islam et al., 2024). For foodborne disease surveillance, prevention, and control, pathogenic organism identification is extremely important. In a previous study, Duijkeren et al. (2003) have shown that antimicrobial resistance in humans and animals is crucial for (a) detecting changes in resistance patterns, (b) implementing control measures regarding the use of antimicrobial agents, and (c) preventing the widespread transmission of multidrug-resistant strains of bacteria. Due to a lack of awareness, understanding, and compliance with food regulations, food contamination and food-borne illnesses are quite prevalent in Bangladesh. Additionally, S. aureus is a pathogen that can be transmitted from animals to humans, leading to infections in people and causing a variety of symptoms. Food poisoning is thought to be caused by S. aureus propensity to develop biofilm on food contact surfaces. S. aureus that forms biofilms is a severe health threat to people. There is currently a lack of reports regarding the prevalence of biofilm-forming S. aureus in various fast food samples from Bangladesh (Ballah et al., 2022). In contrast, E. coli is a widespread and typically harmless bacterium found in the human intestines. Nonetheless, five distinct types of E. coli have been identified as producing diarrhea in warm-blooded animals and people. Different types of E. coli are known for their effects on the digestive system. Some of these are ETEC, EPEC, EAEC, EIEC, and EHEC. The second group comprises STX-ECs, or E. coli that produce Shiga toxin (Ayamah et al., 2021). The findings indicate that fast food sold on the streets in this area is contaminated with foodborne bacteria, which could lead to significant health issues such as Diarrhea, vomiting, gastrointestinal infections. The antimicrobial susceptibility test results indicated that 70–100% of isolates from two organisms exhibited resistance to ampicillin, tetracycline, and erythromycin, complicating the treatment of infections. The nuc gene containing S. aureus showing 100% resistance against Cefoxitin and Oxacillin, and our research aligns entirely with the previous study conducted by Urmi et al. (2021), which observed the highest resistance rates for ampicillin, cephalexin, and vancomycin against S. aureus. Additionally, in previously published studies in Bangladesh, Habib (2016) revealed that Erythromycin and ampicillin were 100% resistant and Ceftazidime and Tetracycline were 80% resistant to E. coli, which support our research. Li et al. (2022) investigated their study erythromycin was found resistant 84.2%, Penicillin (100%), tetracycline (47.2%). Moreover, from this study we investigated that multi-drug-resistant organisms present a significant challenge to human survival and the ongoing existence concerning bacterial infections and diseases. The findings from this study indicate that samples of street vending fast food are contaminated with a range of bacteria, many of which exhibit resistance to commonly used antibiotics, thus posing significant risks and public health concerns. The high percentage of resistance to many antibiotics indicates that E. coli isolates are becoming increasingly resistant to antibiotics. The rise of multidrug resistance in E. coli underscores the importance of enhancing public awareness, educating healthcare professionals, and implementing effective measures to reduce the indiscriminate use of antibiotics. Policymakers should strengthen regulations on antibiotic use in agriculture and enforce food safety standards, while healthcare providers must prioritize infection control and antimicrobial stewardship. The general public should practice proper hygiene, safe food handling, and support sustainable practices to reduce the spread of MDR bacteria. To the best of our knowledge, due to research duration and less sample size, we could not point out the actual prevalence of E. coli and S. aureus from different fast food in Gazipur city. We will do another research on whole Bangladesh and find a significant result that helps to increase public awareness about taking fast food from local shops and restaurants.
5. Conclusions
The investigation yielded valuable information regarding the bacteriological quality of fast food samples and the presence of pathogenic bacterial isolates in Fuchka, Burger, Sandwich, and Chicken Roll, as served in various hotels and restaurants throughout Gazipur city. The findings of our study indicate that certain raw fast food items provided in the selected hotels and restaurants within the locality exhibit bacterial loads that surpass permissible levels. Notable to this study is the identification of potentially harmful bacteria, including E. coli and S. aureus that present significant dangers to consumer health. An increased proportion of multidrug-resistant isolates may pose major risks to handlers, consumers, and the environment. Moreover, most restaurant vendors lack training in food safety and foodborne disease management, resulting in inadequate personal hygiene, improper handling practices, extended storage of food at ambient temperatures, and the sale of leftover food the next day. Therefore, it is essential to implement educational facilities and food safety training programs for all restaurant vendors, which could reduce the risk of infection and ensure the delivery of safe food. In addition, customers should be informed about the unsanitary nature of fast food and urge food sellers to abide by food safety laws. The primary outcome should involve regular monitoring by the health ministry, accompanied by stringent law enforcement, to improve the hygienic practices of fast food establishments.
Acknowledgements
The authors want to give special thanks to the Department of Microbiology, Gono Bishwabidyalay Savar, Dhaka-1344, Bangladesh, for their laboratory facilities, and also thanks to the restaurant owners for supporting us in giving samples.
Data availability
The raw data are available in corresponding author and ready to submit when ask for it.
Informed consent statement
No informed consent was required to conduct the study.
Conflict of interest
The authors declare no conflict of interest.
Authors’ contribution
Conceptualization: Nusrat Jahan, Md. Shah Alam, Bibi Joynab Maria, Atikur Rahman Titas, Saifullah Mansur. Study conduct and manuscript writing: Zakaria Ahmed Sany, Md. Shah Alam, Bibi Joynab Maria, Nusrat Jahan, Farzana Afroz Rimy. Methodology preparation: Nusrat Jahan, Md. Shah Alam, Md. Aoulad Hosen, Farzana Afroz Rimy. Result interpretation: Ahad Alam, Md. Faishal Ahmed, Md. Khairul Islam, Farzana Afroz Rimy. Table formatting: Jahurul Islam, Md. Zahidul Ismam Fahim. Figure formatting: Zakaria Ahmed Sany, Ahad Alam. Final manuscript draft, reviewing: Md. Aoulad Hosen, Zakaria Ahmed Sany, Md. Shah Alam, Bibi Joynab Maria. Final reviewed and revised: Md. Aoulad hosen, Md. Khairul Islam, Saifullah Mansur, Md. Shah Alam, and Farzana Afroz Rimy. All authors critically reviewed the manuscript and agreed to submit final version of the article.



